Day 1: What is Perl Data Language?

If you have a bunch of numerical data that you need crunched fast, you need PDL !
PDL stores its values in a "vectorized data structure" which is compact in memory,
usually in doubles and pre-declared sizes.
This allows for fast traversal and manipulation.
The underlying code is written in C for speed, with access to the internal structure
for those who feel the need to tinker.
A PDL object is sometimes refered to as an ndarray (N-dimensional array) to conform with usage in other languages. Simply put, PDL gives you the ability to process large chunks of data at once.
Getting Started
If you're starting from scratch, the first thing to do is to install PDL
(and if you're going for the latest version, my personal preference is the
cpanminus tool to install it with using cpanm in place of cpan).
Once installed, run the perldl command to get the PDL shell so we can start playing
pdl> $x = ones(5) # a 1D array of 5 ones
pdl> p $x
[1 1 1 1 1]
pdl> $y = 5 * $x + sequence(5) # multiply every element of $x by 5 and add [0 1 2 3 4]
pdl> p $y
[5 6 7 8 9]
pdl> $z = sequence(2,2) # a 2x2 matrix
pdl> p $z
[
[0 1]
[2 3]
]
Let me unpack a bit of that for you ...
Data In
You can make a PDL object (or NDArray) out of any sensible perl array like
$vector = pdl( 0, 1, 2, 3, 4 );
$matrix = pdl( [1..3],[4..6]);
There are also more helper functions than just ones and sequence for quickly getting started.
The functions zeroes, random, nan and inf do what you think they might.
For fun, try out xvals(3,3,3), yvals(3,3,3) and zvals(3,3,3) to see what they do.
You can also read in data from files in a number of formats, like CSV, HDF, Matlab and various image formats.
For the astronomers out there, the rfits command for FITS images
$image = rfits $file;
will also give you access to the FITS header data recorded when the image was taken.
Data manipulation
Did you notice the lack of looping when we defined $y above?
PDL tries hard to Do What You Mean with NDArray operations and usually takes care of the looping for you.
So 5 * $x multiplies every element by 5 and if two NDArrays are the same size,
it will operate on each element at the same index,
so $c = $a + $b is the same as cijk = aijk + bijk
for all i, j and k.
Because it does the obvious thing, it might catch you out when your Algebra brain thinks about multiplying matrices or dot products. I'll get to that in another blog post.
Features
Slicing, Dicing and Indexing
People always want to do strange things to data, the same way otherwise normal people drag perfectly happy trees indoors to decorate when the nights get cold and dark. You want to carve up your data like a Wild Haggis and PDL lets you extract or operate on specific indices (or not those indices) of your ndarrays.
You can move stuff around with mv(), exchange two dimensions with xchg(), shift elements along with rotate()
or just reorder() all the dimensions at once.
Here's an example of creating a circular mask to remove the bright bulge of a galaxy in order to calculate the total luminosity of its spiral arms.
$galaxy = rfits "m51.fits";
$radial = rvals $galaxy; # values increase radially
$mask = $radial<50; # a solid circle around the centre, with radius of 50
$disk = $galaxy * (1 - $mask);
$luminosity = sum $disk;
which would look like
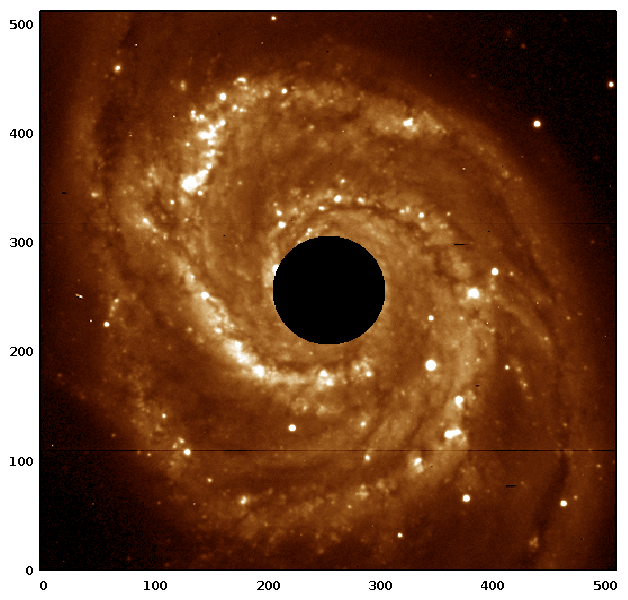
See the PDL Book for a complete discussion on doing photometry with masks.
Don't like the slicing syntax? Try PDL::NiceSlice
Bad Values
Do you have missing or unreliable data? The first two ideas you think of how to handle this problem are probably wrong. Using Bad Values is the right way to mark points that shouldn't be used in calculations. Most PDL functions are aware of Bad Values and will propagate them correctly for you. I will go further into this topic in a later post.
REPL included
As seen above,
PDL comes with a built-in Read-Eval-Print Loop
(the perldl shell) for you to explore the language as well as your data.
Every statement is evaluated as soon has you hit Enter and the shell has a few shortcuts,
such as p to pretty print a data structure soon to be joined by x which will Data::Dumper your data.
And so much more!
That's not even the tip of the iceberg. Also available are:
- Even More Statistical tools which include
- The Simplex optimization algorithm
- Nonlinear least squares fitting routines
- An interface to the Fastest Fourier Transform in the West v3
- An interface to some routines of the BLAS and LAPACK library
to name a few.
PDL Culture
The Early Days of PDL
PDL has been around a long time, released before Numpy. But whereas, pythonistas load numpy at the drop of a hat, Perl has a lot of alternatives before bringing out the big guns. My personal view is that when PDL was released in 1996, a bunch of people jumped on it, got it into a state where you could do everything with it and when it hit version 2.4, they kinda moved on to their own stuff. They'd spent too much time getting stuff done and not enough time telling everybody about it. To be fair, at the time the modes of information transfer were maillists and websites which are harder to stumble across. YouTube hadn't even got started by then, let alone a growing repository of tutorials at your fingertips.
Current state of PDL
Since 2019, the project has been driven by a Release Early, Release Often mantra and is currently at 2.095 (Nov 2024). Ed and the PDL Porters have been smashing bugs and improving performance.
Questions following this talk at the London Perl Workshop start with Is it like Numpy? at 9:40. The short answer is Yes, but better ;)
Oh, and it also plays well with (meaning it has bindings for) GNU Scientific Library, OpenCV, OpenGL, LAPACK and Gnuplot, amongst others. And now there's an easy install of PDL on MacOS. What's not to like!
A Few Resources
Here's a few webpages to keep you busy while you're waiting for the next Advent post.
- Main website https://pdl.perl.org/
- The PDL Book
- 100 PDL Exercises with some answers still left to find
Happy Number Crunching!
The Old Rodney, The Wharfage, Ironbridge - red door and faded old pub signs" by ell brown is licensed under CC BY 2.0
Boyd Duffee
Boyd has wanted to learn PDL for many years and realizing that dream is bringing him joy. He has done mad things to Complex Networks with NLP and is moving on to DSP and Time Series Analysis. He's interested in Data Science, Complex Networks and walks in the woods.